Use Case: Inspecting and Removing Bias from CelebA¶
In this tutorial, we show how to wrap a ResNet model with Neo wrapper, and based on the results we can analyze the bias in the dataset and model, then resample the dataset with a debiased scheme.
CelebFaces Attributes Dataset (CelebA) is a large-scale face attributes dataset with more than 200K celebrity images, each with 40 binary attribute annotations.
Step 1: Initial Setup¶
Download CelebA dataset and model weights¶
Download the dataset here. Unzip the data into folder “…/CelebA”
Download the weights here and place the .pth file in the folder where this notebook is located
The weights used for the base model were trained from the github repo: https://github.com/blingenf/celeba-baselines, using the dataset class for CelebA, which is
celeba.py.
You should setup your dataset structure as described here, using the aligned images.
Import Dependencies¶
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.utils.data import Dataset, DataLoader
from torchvision import models, datasets, transforms
import torchvision.transforms.functional as TF
from urllib.request import urlopen
from glob import glob
from celeba import CelebA
import sys
import time
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from matplotlib.offsetbox import OffsetImage, AnnotationBbox
import matplotlib.gridspec as gridspec
import os
from sklearn.manifold import TSNE
CELEBA_DIR = "CelebA"
Define and initialize the model¶
The base model is ResNet18, we simply need to change the last layer from 1000 outputs to 40, then load the trained weights.
import warnings
warnings.filterwarnings("ignore")
class AttributeNN(nn.Module):
"""Base network for learning representations. Just a wrapper for
ResNet18 which maps the last layer to 40 outputs instead of the
1000 used for ImageNet classification.
"""
def __init__(self, n_labels, pretrain=False):
super().__init__()
self.resnet = models.resnet18(pretrained=pretrain)
self.fc_in_feats = self.resnet.fc.in_features
self.resnet.fc = nn.Linear(self.fc_in_feats, n_labels, bias=True)
self.n_labels = n_labels
def forward(self, input):
output = self.resnet(input)
return output
saved_model_path = 'resnet18_multiplicative'
device='cuda'
saved_state_dict = torch.load(saved_model_path)
model = AttributeNN(40)
model = model.to(device)
# Load the updated state_dict into the model
model.load_state_dict(saved_state_dict)
<All keys matched successfully>
Step 2: Wrap with Capsa-Torch and Train Model¶
Here we use the Neo Wrapper which yields vacuity loss. The model wrapped with Neo Wrapper must be trained, here we provide the training code, and also the weights trained with the same setup for you to load.
from capsa_torch import neo
wrapper=neo.Wrapper()
wrapped_model=wrapper(model)
wrapped_model=wrapped_model.to(device)
Define Training Loop¶
def evaluate(network, dataloader):
with torch.no_grad():
for batch, _ in dataloader:
batch = batch.to(device)
output, risk = network(batch, return_risk=True)
def train(network, dataloader, dataloader_val, lr, epochs, device, model_name, patience=5, delta=0):
"""Simplified function for training the Wrapped_AttributeNN model with early stopping."""
optimizer = optim.SGD(network.parameters(),
lr=lr, momentum=0.9, weight_decay=0.0001)
training_losses = []
validation_losses = []
best_val_loss = float('inf')
epochs_no_improve = 0
network.train()
for epoch in range(epochs):
start_time = time.time()
avg_loss = 0
for i, (batch, _) in enumerate(dataloader):
batch = batch.to(device)
optimizer.zero_grad()
output, risk = network(batch, return_risk=True)
# Compute the loss
loss = risk.mean()
loss.backward()
optimizer.step()
avg_loss += loss.item()
avg_loss /= len(dataloader)
training_losses.append(avg_loss)
print(f"Epoch [{epoch + 1}/{epochs}], Training Loss: {avg_loss:.4f}, Time: {time.time() - start_time:.2f}s")
sys.stdout.flush()
#torch.save(network.state_dict(), f"{model_name}_epoch_{epoch + 1}.pth")
network.eval()
with torch.no_grad():
avg_val_loss = 0
for batch, _ in dataloader_val:
batch = batch.to(device)
output, risk = network(batch, return_risk=True)
loss = risk.mean()
avg_val_loss += loss.item()
avg_val_loss /= len(dataloader_val)
validation_losses.append(avg_val_loss)
print(f"Epoch [{epoch + 1}/{epochs}], Validation Loss: {avg_val_loss:.4f}")
sys.stdout.flush()
# Early stopping logic
if avg_val_loss < best_val_loss - delta:
best_val_loss = avg_val_loss
epochs_no_improve = 0
torch.save(network.state_dict(), f"{model_name}_best.pth") # Save the best model
else:
epochs_no_improve += 1
if epochs_no_improve >= patience:
print(f"Early stopping at epoch {epoch + 1}. Best validation loss: {best_val_loss:.4f}")
break
network.train()
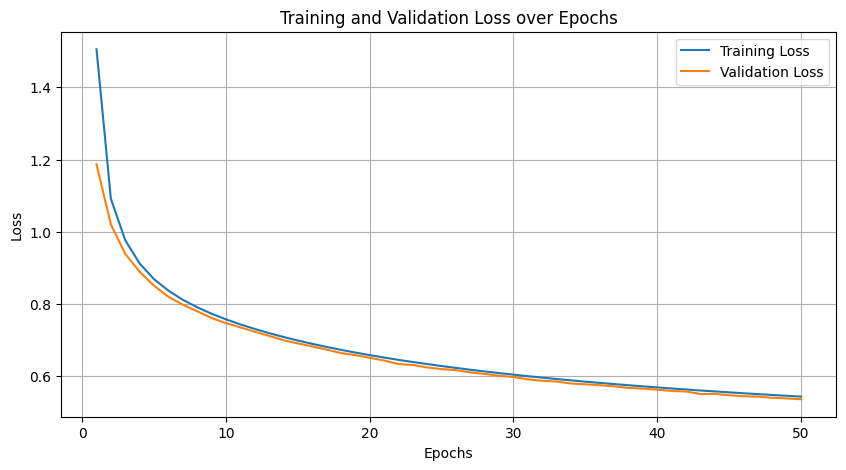
# Plot training and validation losses
plt.figure(figsize=(10, 5))
plt.plot(range(1, epochs + 1), training_losses, label='Training Loss')
plt.plot(range(1, epochs + 1), validation_losses, label='Validation Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.title('Training and Validation Loss over Epochs')
plt.legend()
plt.grid(True)
plt.show()
Load Data and Start Training¶
# Load your dataset
dataset = CelebA(CELEBA_DIR, fold='train', use_transforms=True, normalize=False)
dataloader = DataLoader(dataset, batch_size=32, shuffle=True, num_workers=4)
dataset_val = CelebA(CELEBA_DIR, fold='val', use_transforms=True, normalize=False)
dataloader_val = DataLoader(dataset_val, batch_size=32, shuffle=False, num_workers=4)
dataset_test = CelebA(CELEBA_DIR, fold='test', use_transforms=True, normalize=False)
dataloader_test = DataLoader(dataset_test, batch_size=1, shuffle=False, num_workers=4)
evaluate(wrapped_model, dataloader_val)
# Train the model
train(wrapped_model, dataloader, dataloader_val, lr=0.0001, epochs=50, device='cuda', model_name="Wrapped_AttributeNN")
Epoch [1/50], Training Loss: 1.5056, Time: 227.80s
Epoch [1/50], Validation Loss: 1.1868
Epoch [2/50], Training Loss: 1.0912, Time: 224.38s
Epoch [2/50], Validation Loss: 1.0202
Epoch [3/50], Training Loss: 0.9766, Time: 224.46s
Epoch [3/50], Validation Loss: 0.9390
Epoch [4/50], Training Loss: 0.9120, Time: 223.98s
Epoch [4/50], Validation Loss: 0.8892
Epoch [5/50], Training Loss: 0.8688, Time: 223.50s
Epoch [5/50], Validation Loss: 0.8506
Epoch [6/50], Training Loss: 0.8369, Time: 207.15s
Epoch [6/50], Validation Loss: 0.8201
Epoch [7/50], Training Loss: 0.8117, Time: 202.59s
Epoch [7/50], Validation Loss: 0.7982
Epoch [8/50], Training Loss: 0.7909, Time: 203.23s
Epoch [8/50], Validation Loss: 0.7799
Epoch [9/50], Training Loss: 0.7731, Time: 199.91s
Epoch [9/50], Validation Loss: 0.7612
Epoch [10/50], Training Loss: 0.7576, Time: 198.93s
Epoch [10/50], Validation Loss: 0.7471
Epoch [11/50], Training Loss: 0.7435, Time: 200.88s
Epoch [11/50], Validation Loss: 0.7352
Epoch [12/50], Training Loss: 0.7309, Time: 198.27s
Epoch [12/50], Validation Loss: 0.7231
Epoch [13/50], Training Loss: 0.7193, Time: 197.92s
Epoch [13/50], Validation Loss: 0.7118
Epoch [14/50], Training Loss: 0.7087, Time: 198.96s
Epoch [14/50], Validation Loss: 0.6998
Epoch [15/50], Training Loss: 0.6989, Time: 201.35s
Epoch [15/50], Validation Loss: 0.6908
Epoch [16/50], Training Loss: 0.6897, Time: 199.39s
Epoch [16/50], Validation Loss: 0.6826
Epoch [17/50], Training Loss: 0.6811, Time: 199.39s
Epoch [17/50], Validation Loss: 0.6737
Epoch [18/50], Training Loss: 0.6731, Time: 198.66s
Epoch [18/50], Validation Loss: 0.6641
Epoch [19/50], Training Loss: 0.6655, Time: 199.24s
Epoch [19/50], Validation Loss: 0.6583
Epoch [20/50], Training Loss: 0.6584, Time: 197.32s
Epoch [20/50], Validation Loss: 0.6509
Epoch [21/50], Training Loss: 0.6517, Time: 198.94s
Epoch [21/50], Validation Loss: 0.6435
Epoch [22/50], Training Loss: 0.6453, Time: 199.80s
Epoch [22/50], Validation Loss: 0.6336
Epoch [23/50], Training Loss: 0.6393, Time: 198.46s
Epoch [23/50], Validation Loss: 0.6310
Epoch [24/50], Training Loss: 0.6336, Time: 198.63s
Epoch [24/50], Validation Loss: 0.6242
Epoch [25/50], Training Loss: 0.6281, Time: 200.83s
Epoch [25/50], Validation Loss: 0.6197
Epoch [26/50], Training Loss: 0.6229, Time: 198.24s
Epoch [26/50], Validation Loss: 0.6168
Epoch [27/50], Training Loss: 0.6180, Time: 197.36s
Epoch [27/50], Validation Loss: 0.6104
Epoch [28/50], Training Loss: 0.6132, Time: 199.76s
Epoch [28/50], Validation Loss: 0.6063
Epoch [29/50], Training Loss: 0.6086, Time: 207.99s
Epoch [29/50], Validation Loss: 0.6010
Epoch [30/50], Training Loss: 0.6043, Time: 199.93s
Epoch [30/50], Validation Loss: 0.5978
Epoch [31/50], Training Loss: 0.6001, Time: 200.21s
Epoch [31/50], Validation Loss: 0.5915
Epoch [32/50], Training Loss: 0.5961, Time: 204.00s
Epoch [32/50], Validation Loss: 0.5873
Epoch [33/50], Training Loss: 0.5922, Time: 200.01s
Epoch [33/50], Validation Loss: 0.5854
Epoch [34/50], Training Loss: 0.5885, Time: 200.47s
Epoch [34/50], Validation Loss: 0.5797
Epoch [35/50], Training Loss: 0.5849, Time: 198.26s
Epoch [35/50], Validation Loss: 0.5775
Epoch [36/50], Training Loss: 0.5814, Time: 199.87s
Epoch [36/50], Validation Loss: 0.5751
Epoch [37/50], Training Loss: 0.5782, Time: 198.67s
Epoch [37/50], Validation Loss: 0.5719
Epoch [38/50], Training Loss: 0.5750, Time: 201.08s
Epoch [38/50], Validation Loss: 0.5676
Epoch [39/50], Training Loss: 0.5719, Time: 199.91s
Epoch [39/50], Validation Loss: 0.5657
Epoch [40/50], Training Loss: 0.5690, Time: 199.47s
Epoch [40/50], Validation Loss: 0.5628
Epoch [41/50], Training Loss: 0.5660, Time: 199.95s
Epoch [41/50], Validation Loss: 0.5590
Epoch [42/50], Training Loss: 0.5632, Time: 198.68s
Epoch [42/50], Validation Loss: 0.5577
Epoch [43/50], Training Loss: 0.5604, Time: 198.22s
Epoch [43/50], Validation Loss: 0.5507
Epoch [44/50], Training Loss: 0.5579, Time: 200.46s
Epoch [44/50], Validation Loss: 0.5510
Epoch [45/50], Training Loss: 0.5553, Time: 198.73s
Epoch [45/50], Validation Loss: 0.5473
Epoch [46/50], Training Loss: 0.5528, Time: 197.38s
Epoch [46/50], Validation Loss: 0.5446
Epoch [47/50], Training Loss: 0.5504, Time: 197.71s
Epoch [47/50], Validation Loss: 0.5435
Epoch [48/50], Training Loss: 0.5480, Time: 201.10s
Epoch [48/50], Validation Loss: 0.5403
Epoch [49/50], Training Loss: 0.5457, Time: 201.57s
Epoch [49/50], Validation Loss: 0.5385
Epoch [50/50], Training Loss: 0.5434, Time: 199.72s
Epoch [50/50], Validation Loss: 0.5363
(Optional) Load Saved Weights¶
Instead of training the model in the notebook, one can also download the weights trained with the same setup as above from this link. Then load the weights from this file.
wrapped_model.load_state_dict(torch.load("Wrapped_AttributeNN_best.pth"))
wrapped_model.to(device)
Step 3: Analyze Bias in Dataset¶
Vacuity uncertainty arises from a lack of evidence. A higher value indicates that the data point is less likely to have been drawn from the same distribution as the training data. The Neo wrapper outputs a loss reflecting vacuity uncertainty, but this loss is unnormalized. If a data point has a high vacuity loss but is expected to be common in the real world—or conversely, has a low vacuity loss but is uncommon—this suggests bias in the dataset or the model. We can analyze such bias in the CelebA dataset using vacuity loss as a metric.
Evaluate Testing Dataset¶
losses = []
image_ids = []
true_labels=[]
output_vectors = []
intermediate_vectors=[]
test_labels = []
wrapped_model.eval()
with torch.no_grad():
for image_idx, (images, labels) in enumerate(dataloader_test):
images, labels = images.to('cuda'), labels.to('cuda')
outputs, risk=wrapped_model(images, return_risk=True)
loss = risk.mean()
output_vectors.append(outputs.cpu().numpy())
losses.append(loss.cpu())
image_ids.append(image_idx)
true_labels.append(labels)
test_labels.append(labels.cpu().numpy())
output_vectors = np.concatenate(output_vectors)
test_labels = np.concatenate(test_labels)
losses=np.array(losses)
# Identify the top 10 images with the highest loss
top_10_indices = sorted(range(len(losses)), key=lambda i: losses[i], reverse=True)[:10]
# Output the images and their IDs
top_10_images = [dataset_test[i][0] for i in top_10_indices]
top_10_image_ids = [image_ids[i] for i in top_10_indices]
top_10_image_true_labels = [true_labels[i] for i in top_10_indices]
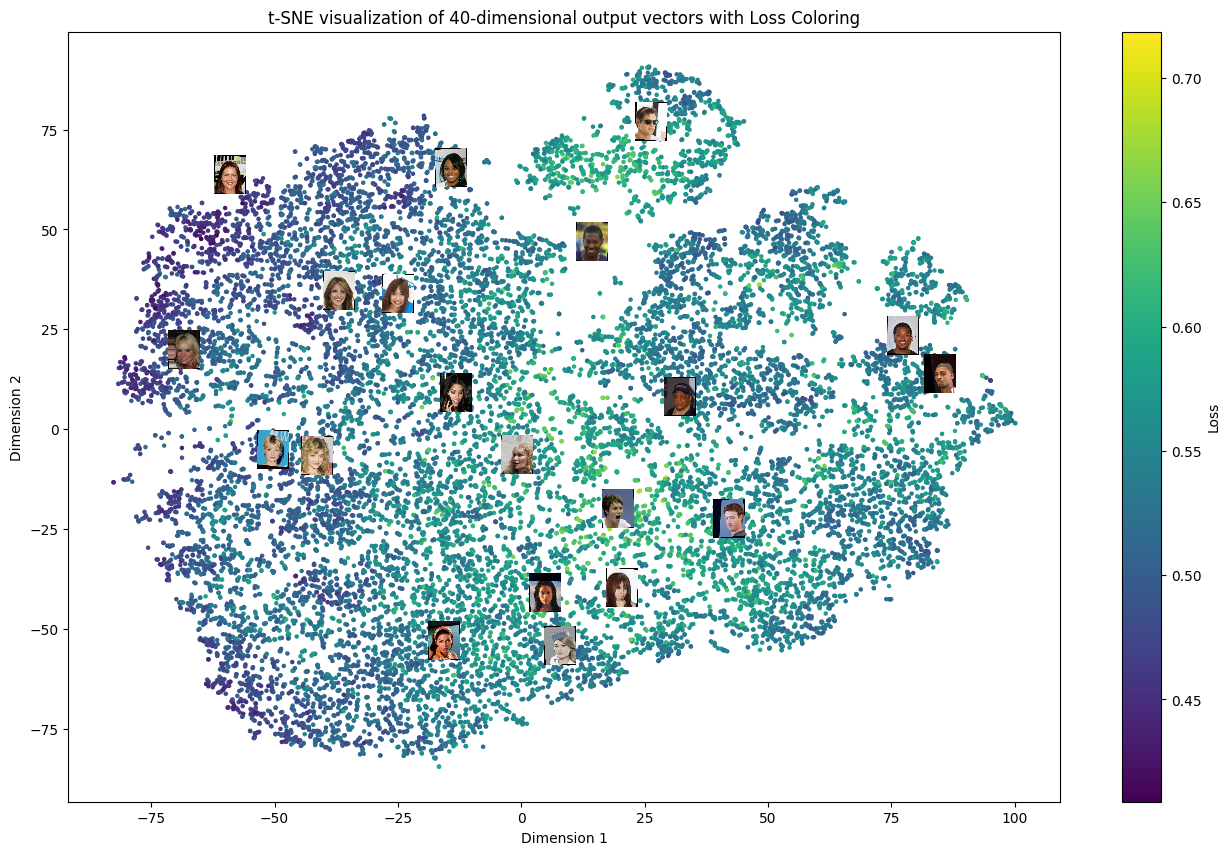
Plot Losses in t-SNE Space¶
To visualize the statistical properties of the model’s outputs, we apply t-SNE to the final outputs of the wrapped model, projecting them into a two-dimensional space. In the resulting figure, we use vacuity loss values as a color map over the t-SNE feature space; brighter colors indicate higher vacuity loss, which corresponds to less typical images in the dataset. Additionally, we randomly select several data points and display their corresponding images near their positions on the plot.
# Apply t-SNE to reduce to 2 dimensions
tsne = TSNE(n_components=2, random_state=42)
tsne_result = tsne.fit_transform(output_vectors)
num_images = len(dataset_test)
# Randomly sample 20 indices from all images
np.random.seed(42) # For reproducibility
sampled_indices = np.random.choice(num_images, 20, replace=False)
norm = plt.Normalize(vmin=losses.min(), vmax=losses.max())
cmap = cm.viridis
# Plot the t-SNE results with loss-based coloring
plt.figure(figsize=(16, 10))
sc = plt.scatter(tsne_result[:, 0], tsne_result[:, 1], c=losses, cmap=cmap, s=5, norm=norm)
plt.colorbar(sc, label='Loss')
# Function to add small images to the plot
def add_image(ax, image, xy):
im = OffsetImage(image, zoom=0.1)
ab = AnnotationBbox(im, xy, xybox=(20, 20), frameon=False, boxcoords="offset points", pad=0.3)
ax.add_artist(ab)
# Add images at the specified indices
ax = plt.gca()
for idx in sampled_indices:
add_image(ax, dataset_test[idx][0].cpu().numpy().transpose(1, 2, 0), tsne_result[idx])
plt.title('t-SNE visualization of 40-dimensional output vectors with Loss Coloring')
plt.xlabel('Dimension 1')
plt.ylabel('Dimension 2')
plt.show()
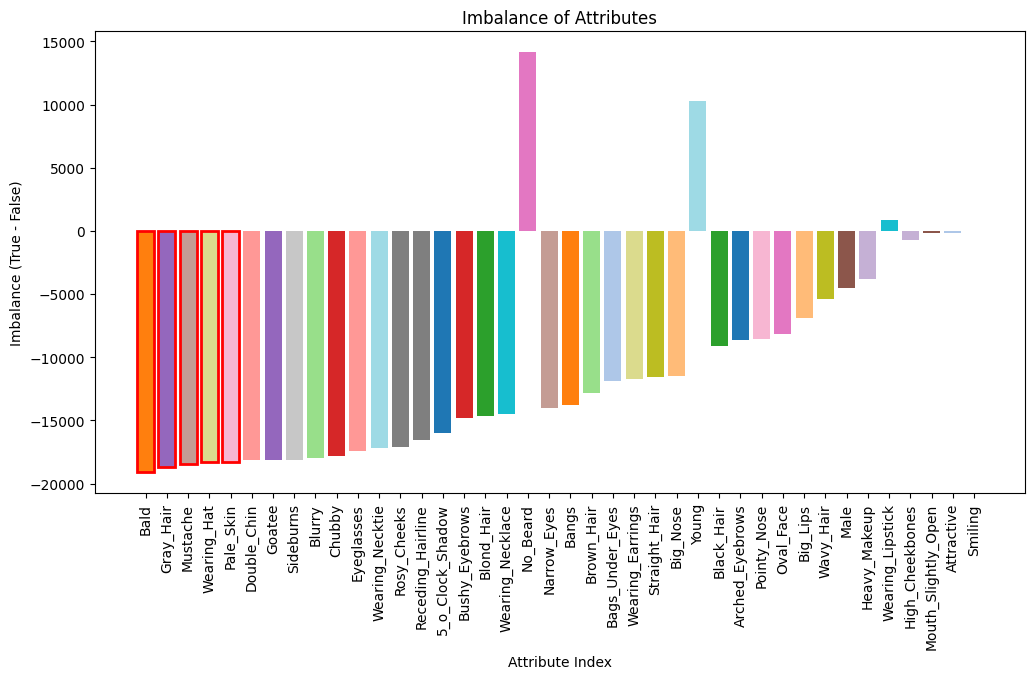
Plot Attribute Imbalance¶
The dataset comprises 40 binary attributes, each labeled as either true or false. In the bar plot below, we display the imbalance for each attribute in the testing dataset, calculated as the number of true labels minus the number of false labels. The attributes are arranged in decreasing order of imbalance, so those with the highest imbalance appear first.
def load_attribute_names(file_path):
with open(file_path, 'r') as f:
lines = f.readlines()
# The second line contains the attribute names
attribute_names = lines[1].strip().split()
return attribute_names
# Define the function to get the attribute values for a specific image index
def get_image_attributes(dataset, index):
image, attributes = dataset[index]
return image, attributes.numpy()
attr_file_path = os.path.join(CELEBA_DIR, 'labels', 'list_attr_celeba.txt')
attribute_names = load_attribute_names(attr_file_path)
# Calculate the imbalance for each attribute
imbalances = []
true_counts = []
false_counts = []
imbalances_ratio=[]
for attribute_index in range(40):
true_count = np.sum(test_labels[:, attribute_index] == 1)
false_count = np.sum(test_labels[:, attribute_index] == 0)
imbalance = true_count - false_count
imbalances_ratio.append((true_count/(true_count+false_count), attribute_index, attribute_names[attribute_index]))
imbalances.append((imbalance, attribute_index, attribute_names[attribute_index]))
# Sort by imbalance and get the top 5 most unbalanced attributes
imbalances.sort(reverse=True, key=lambda x: abs(x[0]))
top_5_unbalanced = [idx for _, idx, _ in imbalances[:5]]
# Sort by imbalance
sorted_imbalances = [imb for imb, idx,_ in imbalances]
sorted_attributes = [idx for imb, _,idx in imbalances]
sorted_attributes_index = [idx for imb, idx,_ in imbalances]
# Plot the bar chart
plt.figure(figsize=(12, 6))
colors = cm.get_cmap('tab20', 40)
bars = plt.bar(range(40), sorted_imbalances, tick_label=sorted_attributes, color=[colors(i) for i in sorted_attributes_index])
# Highlight the top 5 most imbalanced attributes with red outlines
for i in range(5):
bars[i].set_edgecolor('red')
bars[i].set_linewidth(2)
plt.xticks(rotation=90) # Rotate labels to be vertical
plt.xlabel('Attribute Index')
plt.ylabel('Imbalance (True - False)')
plt.title('Imbalance of Attributes')
plt.show()
The code block below processes the testing results in preparation for the figure plotted in the next code block. We start by filtering the vacuity losses to a smaller range because only a few images have very large vacuity losses outside this range. By focusing on this smaller range that contains almost all the images, we simplify the histogram we will draw. We then divide the vacuity losses into 10 subranges and count the number of images in each subrange for the histogram to be plotted next. The rest of the code identifies, within each vacuity loss range, the attributes whose distributions differ the most from those in the full testing dataset.
# Set the desired value range for the losses
min_value = 0
max_value = 0.7
# Filter the losses to include only those within the specified range
filtered_losses = [loss for loss in losses if min_value <= loss <= max_value]
# Number of bins
num_bins = 10
# Use numpy's histogram function to get counts and bins without plotting
counts, bins = np.histogram(filtered_losses, bins=num_bins)
# Assign each loss to a bin
bin_indices = np.digitize(losses, bins=bins[:-1], right=False) # Bins are 1-indexed
# Initialize a list to hold labels for each bin
labels_per_bin = [[] for _ in range(num_bins)]
# Group labels by their corresponding bin
for idx, bin_idx in enumerate(bin_indices):
bin_idx = bin_idx - 1 # Adjust bin index to start from 0
labels_per_bin[bin_idx].append(true_labels[idx].cpu()[0])
# Calculate how the imbalances for each attribute in each bin differ from the full testing data
top_5_unbalanced_ratio_attribute_per_bin=[]
top_5_unbalanced_ratio_direction_attribute_per_bin=[]
for bin_idx in range(10):
imbalances = []
imbalances_ratio_diffs = []
for attribute_index in range(40):
true_count = np.sum(np.array(labels_per_bin[bin_idx])[:, attribute_index] == 1)
false_count = np.sum(np.array(labels_per_bin[bin_idx])[:, attribute_index] == 0)
imbalances_ratio_diff = abs(true_count/(true_count+false_count)-imbalances_ratio[attribute_index][0])
imbalances_ratio_diff_direction = (true_count/(true_count+false_count)-imbalances_ratio[attribute_index][0])>0
imbalances_ratio_diffs.append((imbalances_ratio_diff, imbalances_ratio_diff_direction, attribute_index, attribute_names[attribute_index]))
imbalances_ratio_diffs.sort(reverse=True, key=lambda x: x[0])
top_5_unbalanced_ratio_diff = [idx for _, _, idx, _ in imbalances_ratio_diffs[:5]]
top_5_unbalanced_ratio_diff_direction = [idx for _, idx, _, _ in imbalances_ratio_diffs[:5]]
top_5_unbalanced_ratio_diff_attribute= [attribute_names[i] for i in top_5_unbalanced_ratio_diff]
top_5_unbalanced_ratio_attribute_per_bin.append(top_5_unbalanced_ratio_diff_attribute)
top_5_unbalanced_ratio_direction_attribute_per_bin.append(top_5_unbalanced_ratio_diff_direction)
Associate Vacuity with Presence/Absence of Attributes¶
In the top figure below, we present a histogram of the vacuity losses to show their frequency distribution. For each bin in this histogram, we randomly sample four images and display them in the bottom figure. Above each bin, we indicate the top two attributes whose imbalances differ the most from those of the full dataset. A black font indicates the presence of an attribute, while a red font indicates the absence/inverse of the attribute. This figure demonstrates how the values of vacuity losses relate to specific attributes and provides examples of typical images for each range of vacuity losses.
# Function to get 4 images for a given bin range
def get_images_for_loss_range(bin_range, n_samples=4):
bin_indices = [i for i, loss in enumerate(losses) if bin_range[0] <= loss < bin_range[1]]
if len(bin_indices) < n_samples:
n_samples = len(bin_indices)
sampled_indices = np.random.choice(bin_indices, n_samples, replace=False)
images = [dataset_test[i][0].permute(1, 2, 0).cpu().numpy() for i in sampled_indices] # Convert to (H, W, C)
return images
# Collect 4 images for each bin (within the filtered range)
images_per_bin = []
for i in range(num_bins):
bin_range = (bins[i], bins[i+1])
images = get_images_for_loss_range(bin_range)
images_per_bin.append(images)
# Plot the histogram
plt.figure(figsize=(16, 8))
bin_width = (bins[1] - bins[0]) * 0.6
plt.bar(bins[:-1], counts, width=bin_width, edgecolor='black', align='edge')
# Add vertical dashed lines
for i in range(1, len(bins) - 1):
plt.axvline(x=bins[i] - (bins[1] - bins[0]) * 0.2, color='gray', linestyle='--', linewidth=1)
# Determine the fixed y-coordinate for the additional texts
max_count = max(counts)
y_text_fixed = max_count +1000 # Adjust this value as needed
# Add text annotations
for i in range(len(counts)):
bar_center = (bins[i] + bins[i+1]) / 2
y_base = counts[i] + 0.5
plt.text(bar_center, y_base, f'Bin {i+1}', ha='center', fontsize=10)
texts = top_5_unbalanced_ratio_attribute_per_bin[i]
directions = top_5_unbalanced_ratio_direction_attribute_per_bin[i]
for j in range(2):
text = texts[j]
direction = directions[j]
color = 'black' if direction else 'red'
y_offset = y_text_fixed + j * 500
plt.text(bar_center, y_offset, text, ha='center', fontsize=10, color=color)
plt.ylim(top=y_offset + 500)
plt.xlabel('Loss')
plt.ylabel('Count')
plt.title('Histogram with Text Annotations per Bin')
plt.show()
# Create a figure with 1 row and 10 columns for each 2x2 image grid
fig = plt.figure(figsize=(20, 4))
outer_grid = gridspec.GridSpec(1, 10, figure=fig, wspace=0.05, hspace=0)
for i, imgs in enumerate(images_per_bin):
ax = plt.subplot(outer_grid[i])
inner_grid = gridspec.GridSpecFromSubplotSpec(2, 2, subplot_spec=outer_grid[i], wspace=0, hspace=-0.58)
for j, img in enumerate(imgs):
ax2 = plt.Subplot(fig, inner_grid[j])
ax2.imshow(img)
ax2.axis('off')
ax2.set_xticks([])
ax2.set_yticks([])
fig.add_subplot(ax2)
ax.set_title(f'Bin {i+1}', fontsize=16, y=0.8)
ax.axis('off')
fig.suptitle('4 Images for Each Loss Bin (Filtered Range)', fontsize=16)
plt.tight_layout()
plt.subplots_adjust(wspace=0.05, hspace=0)
plt.show()
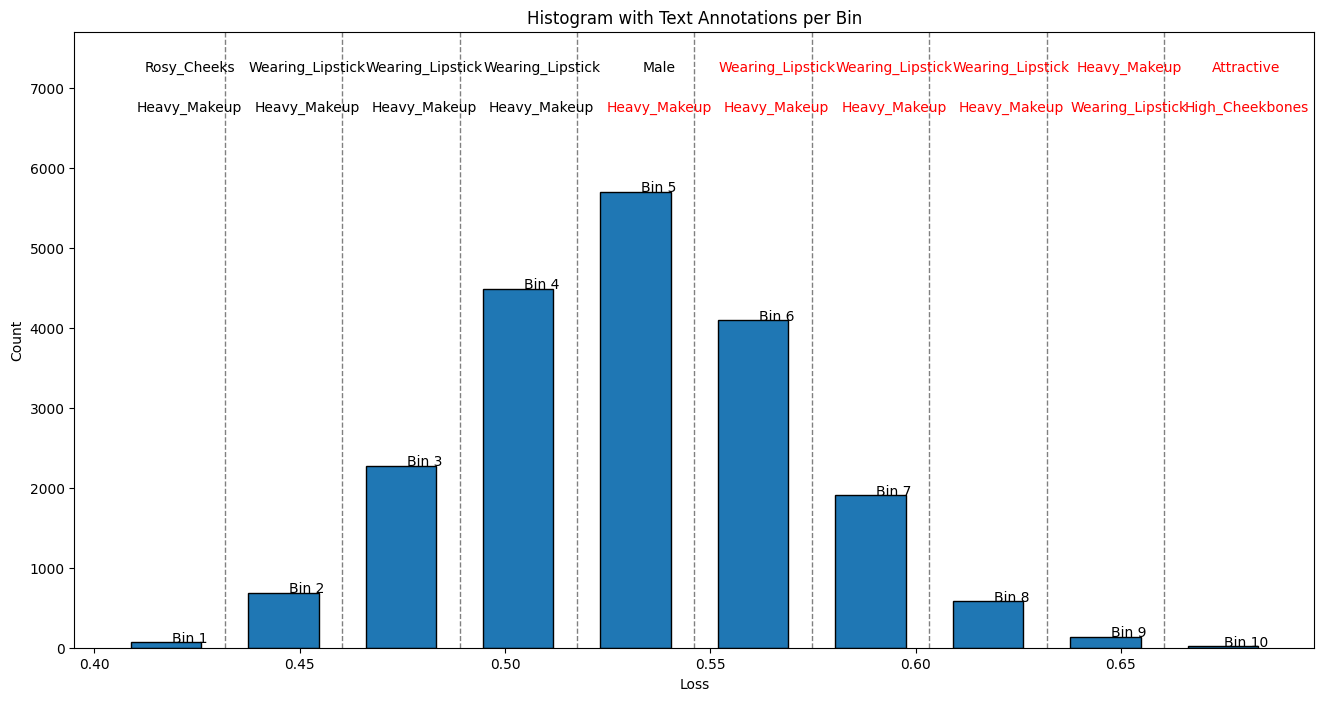
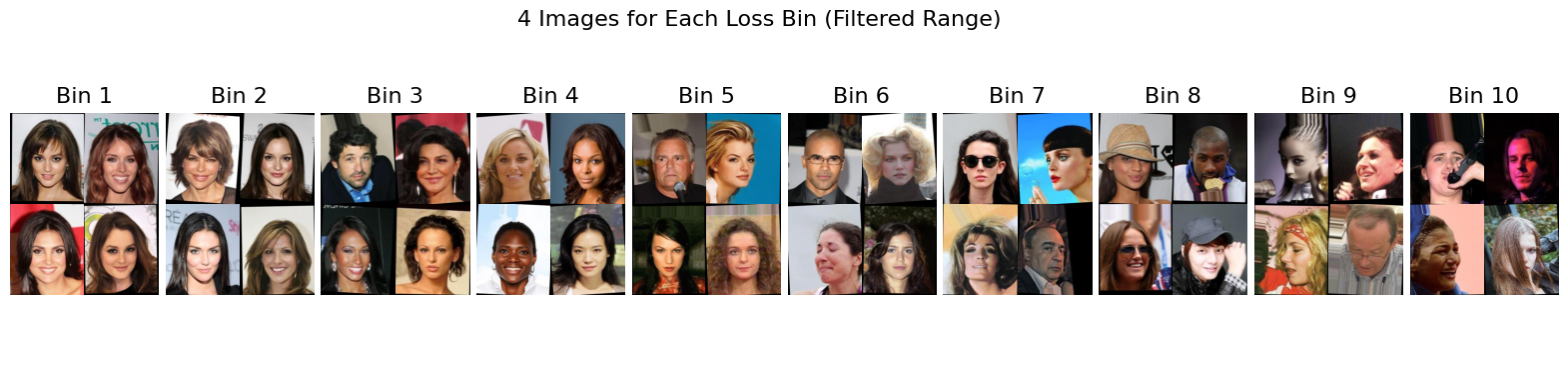
Based on the figure above, we observe that images with the smallest vacuity losses predominantly feature young women. Men also appear frequently among images with low vacuity losses, particularly in Bin 7. As the vacuity loss increases beyond Bin 3, the images exhibit greater diversity, including individuals with darker skin tones, older adults, light or no makeup, and people wearing accessories like glasses or hats.
Step 4: Resample Dataset to Reduce Bias¶
We can resample the testing dataset using sampling schemes that consider the vacuity losses of the images. A straightforward method is to sample images uniformly across bins defined by vacuity loss ranges. This involves two steps: first, we uniformly select a bin corresponding to a specific vacuity loss range; second, we randomly select an image from within that bin. This approach ensures that images from different vacuity loss ranges are equally represented in the resampled dataset.
import numpy as np
import torch
import matplotlib.pyplot as plt
import random
# Define a function to plot images in a 3x3 grid
def plot_images_grid(images, title):
fig, axes = plt.subplots(3, 3, figsize=(8, 8))
fig.suptitle(title, fontsize=16)
for i, ax in enumerate(axes.flat):
ax.imshow(images[i])
ax.axis('off')
plt.tight_layout()
plt.show()
# 1. Uniform Random Sampling
n_samples = 9
uniform_indices = random.sample(range(len(dataset_test)), n_samples)
uniform_sampled_images = [dataset_test[i][0].permute(1, 2, 0) for i in uniform_indices] # dataset_test[i] gives (image, label)
plot_images_grid(uniform_sampled_images, title='Uniformly Sampled Images')
# 2. Uniformly select 9 bins
uniform_bins = random.sample(range(num_bins), n_samples)
uniform_bins_sampled_images=[]
for i in uniform_bins:
bin_range = (bins[i], bins[i+1])
images = get_images_for_loss_range(bin_range,1)
uniform_bins_sampled_images.append(images[0])
plot_images_grid(uniform_bins_sampled_images, title='Weighted Sampled Images Based on Vacuity Loss')


The example above demonstrates that the resampled images, guided by vacuity loss predictions from the wrapped model, exhibit greater diversity. Attributes such as wearing a hat, earrings, or glasses are also treated as factors contributing to the diversity of the dataset since they are included in the training labels.